In the same section
-
Share this page
Image Analysis
- Machine & Deep Learning
-
Biomedical Imaging
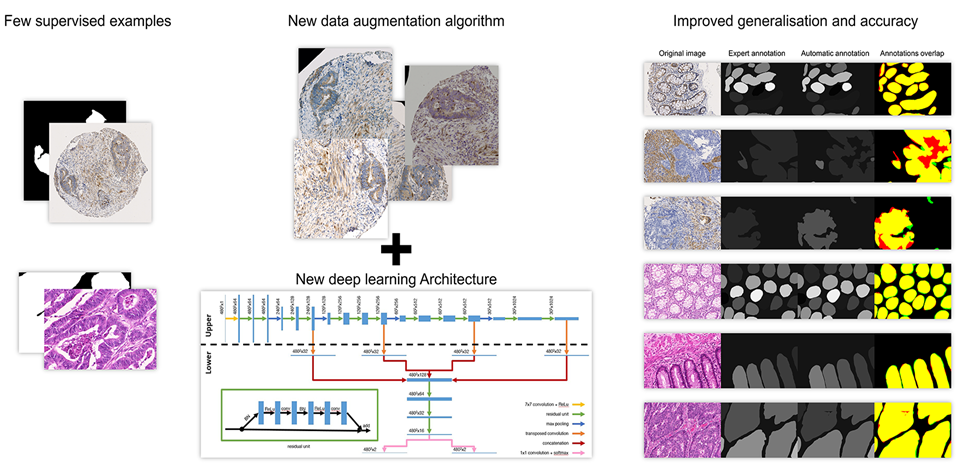
Detection, segmentation and classification are usual tasks in biomedical image analysis. The objects of interest (to detect, segment and/or classify) are often heterogeneous in appearance and therefore difficult to describe using standard image features, which has made deep learning a leading approach to providing effective solutions in this area. However, deep learning algorithms bring their own requirements and challenges with them, such as the need for large amounts of annotated images for model training and validation. Such annotations have to be provided by biomedical experts, but this task is time-consuming, error-prone and expert-dependent, especially if segmentation is required.
Our research aims to develop approaches that are robust to imperfect and incomplete annotations in order to reduce the need for the number and quality of annotations, as well as relevant methods for evaluating models in this context (granted by Walloon Region, FEDER and ULB).
Our fields of application concern :- Whole slide imaging and characterization of tissue-based biomarkers in digital pathology
- Weak supervised in-vitro phase contrast cell tracking : The project aims to provide an object tracking system based on 2D sequences of phase contrast image of cell. The originality of this project is to rely on a very small supervised set of images, and a diverse data augmentation.
Other fields
Document analysis
Research in the area of document analysis at LISA addresses questions concerning the analysis of complex schemes and symbols for recognition and similarity evaluation tasks, involving both structure and semantic analysis from 2D documents.
Applications related to the document analysis:- Piping & Instrumentation diagram (P&ID) analysis; P&ID is a an articulate drawing of a processing plan, including basic symbols (e.g. alphanumeric characters) and domain-specific symbols (e.g. related to railways, industrial piping or bio-pharmaceutical production plant). Geometrical Deep learning is used for complex graph comparison. This research is supported by Innoviris (in collaboration with the industry).
- Detection of trademark infringement using evaluation of intellectual property relevant similarities between logos; Deep learning is used both on the image side for image/image comparison but also for extracting semantic content from text of law associated with court decisions. The project is supported by an ARC project in collaboration with the Juris Lab of the ULB.
- Digitization and quality improvement of old seismic signal records on paper; the aim is to make the resulting historical data FAIR (Findability, Accessibility, Interoperability, and Reusable). Deep learning is used for image filtering and ambiguity resolution (line crossing, stains etc), this research is supported by Belspo and is done in collaboration with the Royal Observatory of Belgium.
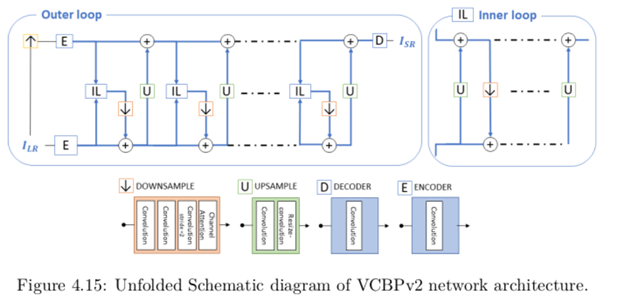
Image super-resolution and image fusion
As the image sensors are becoming cheaper with an increasing resolution, some wavelengths remain more restricted, in particular thermal sensors that remain expensive with respect to their resolution. In that context, we developed an image superresolution algorithm that combines several imaging modalities leveraging the versatility of the deep neural networks and their ability to generale rich and coherent content. Another “beyond the visible spectrum” application consists of generating a RGB visible image from a thermal image alone, extending the night vision possibility.
In collaboration with the VUB (ETRO), we extend this approach to 2D images generated by arrays of microphones; these acoustics images can also have their resolution computationally augmented using these deep neural networks architecture.
Example of network architecture used for super-resolution
Example of super-resolution result obtained on a RGB image (compared with simple bicubic interpolation)
Thermal image colorization (thermal / night rgb / synthetic rgb daylight)
Some publicationsExample: journal 2015 (finds the intersection of the two terms)
2021
?XCycles Backprojection Acoustic Super-Resolution
Almasri, Feras, Vandendriessche, Jurgen, Segers, Laurent, da Silva, Bruno, Braeken, An, Steenhaut, Kris, Touhafi, Abdellah, and Debeir, OlivierSensors, 21, (10), pages 3453, 2021. DOI: 10.3390/s21103453.@article{info:hdl:2013/323448, author={Almasri, Feras and Vandendriessche, Jurgen and Segers, Laurent and da Silva, Bruno and Braeken, An and Steenhaut, Kris and Touhafi, Abdellah and Debeir, Olivier}, title={XCycles Backprojection Acoustic Super-Resolution}, journal={Sensors}, year={2021}, volume={21}, number={10}, pages={3453}, note={Language of publication: en}, note={DOI: 10.3390/s21103453}, abstract={The computer vision community has paid much attention to the development of visible image super-resolution (SR) using deep neural networks (DNNs) and has achieved impressive results. The advancement of non-visible light sensors, such as acoustic imaging sensors, has attracted much attention, as they allow people to visualize the intensity of sound waves beyond the visible spectrum. However, because of the limitations imposed on acquiring acoustic data, new methods for improving the resolution of the acoustic images are necessary. At this time, there is no acoustic imaging dataset designed for the SR problem. This work proposed a novel backprojection model architecture for the acoustic image super-resolution problem, together with Acoustic Map Imaging VUB-ULB Dataset (AMIVU). The dataset provides large simulated and real captured images at different resolutions. The proposed XCycles BackProjection model (XCBP), in contrast to the feedforward model approach, fully uses the iterative correction procedure in each cycle to reconstruct the residual error correction for the encoded features in both low- and high-resolution space. The proposed approach was evaluated on the dataset and showed high outperformance compared to the classical interpolation operators and to the recent feedforward state-of-the-art models. It also contributed to a drastically reduced sub-sampling error produced during the data acquisition.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/323448}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/323448/1/doi\_307092.pdf}, }journal
Processing multi-expert annotations in digital pathology: A study of the Gleason2019 challenge
Foucart, Adrien, Debeir, Olivier, and Decaestecker, ChristineIn Proceedings of SPIE - The International Society for Optical Engineering, 12088, 2021. DOI: 10.1117/12.2604307.@inproceedings{info:hdl:2013/339072, author={Foucart, Adrien and Debeir, Olivier and Decaestecker, Christine}, title={Processing multi-expert annotations in digital pathology: A study of the Gleason2019 challenge}, booktitle={Proceedings of SPIE - The International Society for Optical Engineering}, year={2021}, volume={12088}, eprint={120880X}, note={SCOPUS: cp.p}, note={Language of publication: en}, note={DOI: 10.1117/12.2604307}, abstract={Deep learning algorithms rely on large amounts of annotations for learning and testing. In digital pathology, a ground truth is rarely available, and many tasks show large inter-expert disagreement. Using the Gleason2019 dataset, we analyse how the choices we make in getting the ground truth from multiple experts may affect the results and the conclusions we could make from challenges and benchmarks. We show that using undocumented consensus methods, as is often done, reduces our ability to properly analyse challenge results. We also show that taking into account each expert's annotations enriches discussions on results and is more in line with the clinical reality and complexity of the application.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/339072}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/339072/3/FOUCART21-Gleason2019.pdf}, }conference, workshop
2020
?Thermal Image Super-Resolution Challenge - PBVS 2020
Rivadeneira, Rafael R.E., Sappa, A.D., Vintimilla, Boris B.X., Guo, Lin, Hou, Jiankun, Mehri, Armin, Ardakani, Parichehr Behjati, Patel, Heena, Chudasama, Vishal, Prajapati, Kalpesh, Upla, Kishor K.P., Ramachandra, Raghavendra, Raja, Kiran, Busch, Christoph, Almasri, Feras, Debeir, Olivier, Nathan, Sabari, Kansal, Priya, Gutierrez, Nolan, Bardia, Mojra, and Beksi, William W.J.2020. DOI: 10.1109/CVPRW50498.2020.00056.@inproceedings{info:hdl:2013/317693, author={Rivadeneira, Rafael R.E. and Sappa, A.D. and Vintimilla, Boris B.X. and Guo, Lin and Hou, Jiankun and Mehri, Armin and Ardakani, Parichehr Behjati and Patel, Heena and Chudasama, Vishal and Prajapati, Kalpesh and Upla, Kishor K.P. and Ramachandra, Raghavendra and Raja, Kiran and Busch, Christoph and Almasri, Feras and Debeir, Olivier and Nathan, Sabari and Kansal, Priya and Gutierrez, Nolan and Bardia, Mojra and Beksi, William W.J.}, title={Thermal Image Super-Resolution Challenge - PBVS 2020}, year={2020}, note={Language of publication: en}, note={DOI: 10.1109/CVPRW50498.2020.00056}, abstract={This paper summarizes the top contributions to the first challenge on thermal image super-resolution (TISR), which was organized as part of the Perception Beyond the Visible Spectrum (PBVS) 2020 workshop. In this challenge, a novel thermal image dataset is considered together with state- of-the-art approaches evaluated under a common framework. The dataset used in the challenge consists of 1021 thermal images, obtained from three distinct thermal cameras at different resolutions (low-resolution, mid-resolution, and high-resolution), resulting in a total of 3063 thermal images. From each resolution, 951 images are used for training and 50 for testing while the 20 remaining images are used for two proposed evaluations. The first evaluation consists of downsampling the low-resolution, mid-resolution, and high-resolution thermal images by {\?x}2, {\?x}3 and {\?x}4 respectively, and comparing their super-resolution results with the corresponding ground truth images. The second evaluation is comprised of obtaining the {\?x}2 super-resolution from a given mid-resolution thermal image and comparing it with the corresponding semi-registered high- resolution thermal image. Out of 51 registered participants, 6 teams reached the final validation phase.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/317693}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/317693/3/PBVS\_2020\_CVPRW\_2020.pdf}, }conference, workshop
2019
?Strategies to Reduce the Expert Supervision Required for Deep Learning-Based Segmentation of Histopathological Images
Van Eycke, Yves-Remi, Foucart, Adrien, and Decaestecker, ChristineFrontiers in Medicine, 6, 2019. DOI: 10.3389/fmed.2019.00222.@article{info:hdl:2013/296977, author={Van Eycke, Yves-Remi and Foucart, Adrien and Decaestecker, Christine}, title={Strategies to Reduce the Expert Supervision Required for Deep Learning-Based Segmentation of Histopathological Images}, journal={Frontiers in Medicine}, year={2019}, volume={6}, eprint={222}, note={SCOPUS: re.j}, note={Language of publication: en}, note={DOI: 10.3389/fmed.2019.00222}, abstract={The emergence of computational pathology comes with a demand to extract more and more information from each tissue sample. Such information extraction often requires the segmentation of numerous histological objects (e.g., cell nuclei, glands, etc.) in histological slide images, a task for which deep learning algorithms have demonstrated their effectiveness. However, these algorithms require many training examples to be efficient and robust. For this purpose, pathologists must manually segment hundreds or even thousands of objects in histological images, i.e., a long, tedious and potentially biased task. The present paper aims to review strategies that could help provide the very large number of annotated images needed to automate the segmentation of histological images using deep learning. This review identifies and describes four different approaches: the use of immunohistochemical markers as labels, realistic data augmentation, Generative Adversarial Networks (GAN), and transfer learning. In addition, we describe alternative learning strategies that can use imperfect annotations. Adding real data with high-quality annotations to the training set is a safe way to improve the performance of a well configured deep neural network. However, the present review provides new perspectives through the use of artificially generated data and/or imperfect annotations, in addition to transfer learning opportunities.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/296977}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/296977/1/doi\_280621.pdf}, }journal
SNOW: Semi-Supervised, NOisy and/or Weak Data for Deep Learning in Digital Pathology
Foucart, Adrien, Debeir, Olivier, and Decaestecker, ChristineIn 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), IEEE, pages 1869-1872, 2019. Conference: (April 8-11, 2019: Venice, Italy).@inproceedings{info:hdl:2013/286269, author={Foucart, Adrien and Debeir, Olivier and Decaestecker, Christine}, title={SNOW: Semi-Supervised, NOisy and/or Weak Data for Deep Learning in Digital Pathology}, year={2019}, pages={1869-1872}, note={Language of publication: en}, note={DOI: 10.1109/ISBI.2019.8759545}, note={Conference: (April 8-11, 2019: Venice, Italy)}, publisher={IEEE}, booktitle={2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)}, abstract={Digital pathology produces a lot of images. For machine learning applications, these images need to be annotated, which can be complex and time consuming. Therefore, outside of a few benchmark datasets, real-world applications often rely on data with scarce or unreliable annotations. Inthis paper, we quantitatively analyze how different types of perturbations influence the results of a typical deep learning algorithm by artificially weakening the annotations of a benchmark biomedical dataset. We use classical machine learning paradigms (semi-supervised, noisy and weak learning) adapted to deep learning to try to counteract those effects, and analyze the effectiveness of these methods in addressing different types of weakness.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/286269}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/286269/3/Paper\_ISBI2019\_final.pdf}, }conference, workshop
Data augmentation for training deep regression for in vitro cell detection
Debeir, Olivier, and Decaestecker, ChristineIn Fifth International Conference on Advances in Biomedical Engineering (ICABME), pages 1-3, 2019. Conference: International Conference on Advances in Biomedical Engineering (ICABME)(October 17-19, 2019: Lebanon).@inproceedings{info:hdl:2013/298616, author={Debeir, Olivier and Decaestecker, Christine}, title={Data augmentation for training deep regression for in vitro cell detection}, year={2019}, pages={1--3}, note={Language of publication: fr}, note={DOI: 10.1109/ICABME47164.2019.8940275}, note={Conference: International Conference on Advances in Biomedical Engineering (ICABME)(October 17-19, 2019: Lebanon)}, booktitle={Fifth International Conference on Advances in Biomedical Engineering (ICABME)}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/298616}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/298616/3/tracking2019b.pdf}, }conference, workshop
2018
?Segmentation of glandular epithelium in colorectal tumours to automatically compartmentalise IHC biomarker quantification: a deep learning approach
Van Eycke, Yves-Remi, Balsat, Cédric, Verset, Laurine, Debeir, Olivier, Salmon, Isabelle, and Decaestecker, ChristineMedical image analysis, 49, pages 35-45, 2018. DOI: 10.1016/j.media.2018.07.004.@article{info:hdl:2013/273557, author={Van Eycke, Yves-Remi and Balsat, C{\'e}dric and Verset, Laurine and Debeir, Olivier and Salmon, Isabelle and Decaestecker, Christine}, title={Segmentation of glandular epithelium in colorectal tumours to automatically compartmentalise IHC biomarker quantification: a deep learning approach}, journal={Medical image analysis}, year={2018}, volume={49}, pages={35-45}, note={SCOPUS: ar.j}, note={Language of publication: en}, note={DOI: 10.1016/j.media.2018.07.004}, abstract={In this paper, we propose a method for automatically annotating slide images from colorectal tissue samples. Our objective is to segment glandular epithelium in histological images from tissue slides submitted to different staining techniques, including usual haematoxylin-eosin (H\&E) as well as immunohistochemistry (IHC). The proposed method makes use of Deep Learning and is based on a new convolutional network architecture. Our method achieves better performances than the state of the art on the H\&E images of the GlaS challenge contest, whereas it uses only the haematoxylin colour channel extracted by colour deconvolution from the RGB images in order to extend its applicability to IHC. The network only needs to be fine-tuned on a small number of additional examples to be accurate on a new IHC dataset. Our approach also includes a new method of data augmentation to achieve good generalisation when working with different experimental conditions and different IHC markers. We show that our methodology enables to automate the compartmentalisation of the IHC biomarker analysis, results concurring highly with manual annotations.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/273557}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/273557/3/Article\_segmentation\_glandes\_\_Med\_Im\_An\_postprint.pdf}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/273557/6/Manuscript\_3.pdf}, }journal
RGB Guided Thermal Super-Resolution Enhancement
Almasri, Feras, and Debeir, OlivierIn 4th International Conference on Cloud Computing Technologies and Applications (Cloudtech) 2018, IEEE, 2018. Conference: Cloudtech 2018(4: 26-28 November 2018: Brussels, Belgium).@inproceedings{info:hdl:2013/288194, author={Almasri, Feras and Debeir, Olivier}, title={RGB Guided Thermal Super-Resolution Enhancement}, year={2018}, note={Language of publication: fr}, note={DOI: 10.1109/CloudTech.2018.8713356}, note={Conference: Cloudtech 2018(4: 26-28 November 2018: Brussels, Belgium)}, publisher={IEEE}, series={IEEE Xplore}, booktitle={4th International Conference on Cloud Computing Technologies and Applications (Cloudtech) 2018}, abstract={In visual surveillance and security problems, objects can occur in different conditions of illumination and occlusion, therefore thermal images have become a major tool in a large variety of applications. By the reason of their high cost compared to their visual (RGB) counterpart, thermal sensors are used in low-resolution and in low contrast which introduces the necessity to obtain a higher resolution version. In this work, we propose a deep learning model by which to enhance the thermal image resolution guided by RGB images using GAN based model. The results indicate an improvement in resolution enhancement using RGB guided thermal super-resolution models compared to the classical single thermal super-resolution approach.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/288194}, }conference, workshop
- Whole slide imaging and characterization of tissue-based biomarkers in digital pathology
- Other
-
Biomedical Imaging
Cell tracking: Specific image acquisition and analysis software packages for phase-contrast microscopy were developed for the analysis of cell behavior, in terms of motility, division and death. The module ivctrack is available on github.
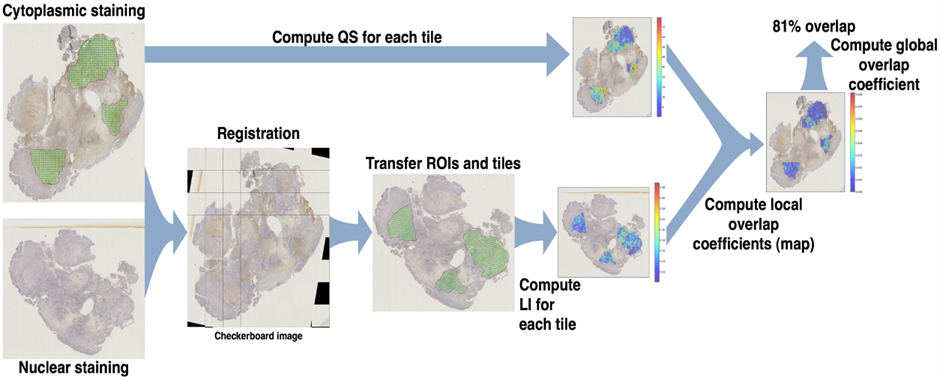
Image registration: Our research aims to develop novel techniques for efficient image registration either at high resolution level and for very large images, or between different imaging modalities with very different resolution levels (e.g., macroscopic vs. microscopic, in vivo vs. histology). This ability is of utmost importance in various medical applications, such as analyses of histopathological biomarker colocalization and therapy effects at the tissue and cell level.
Modeling : Developing and validating a mathematical tumor growth model driven by patient-specific Imaging data to improve surgery and radiotherapy planning
Other fields
Industrial vision:- Various image analysis projects have been carried out in collaboration with our industrial partner Macq-electonics on traffic analysis (vehicle detection, speed estimation, …).
- Arturo project (archive)
People who undergo amputation develop severe pain they locate in the amputated limb: it is called phantom pain. As a therapy, patients can use a mirror to create the illusion of seeing the amputated limb when in reality they are looking at the reflection of the healthy limb. To make the illusion more realistic, we developed an augmented reality system based on images provided by an RGB+d camera. This system constructs a textured mesh to apply a mirror effect and display a realistic 3D image of the patient, in stereovision on a 3D TV or in an immersion helmet, while tracking in real time the central position of the patient's body - Massai Project (archive)
Based on 3D sensors (time of flight, structured light, stereo), we developed a system for the automatic fall detection of elderly people, people in revalidation and people with psychogeriatric disorders in hospitals, rest homes , rest and care homes and service flats. The project was carried out in collaboration with Mintt company.
Some publicationsExample: journal 2015 (finds the intersection of the two terms)
2021
?Initial Condition Assessment for Reaction-Diffusion Glioma Growth Models: A Translational MRI-Histology (In)Validation Study
Martens, Corentin, Lebrun, Laetitia, Decaestecker, Christine, Vandamme, Thomas, Van Eycke, Yves-Remi, Rovai, Antonin, Metens, Thierry, Debeir, Olivier, Goldman, Serge, Salmon, Isabelle, and Van Simaeys, GaëtanTomography, 7, (4), pages 650-674, 2021. DOI: 10.3390/tomography7040055.@article{info:hdl:2013/334520, author={Martens, Corentin and Lebrun, Laetitia and Decaestecker, Christine and Vandamme, Thomas and Van Eycke, Yves-Remi and Rovai, Antonin and Metens, Thierry and Debeir, Olivier and Goldman, Serge and Salmon, Isabelle and Van Simaeys, Ga{\"e}tan}, title={Initial Condition Assessment for Reaction-Diffusion Glioma Growth Models: A Translational MRI-Histology (In)Validation Study}, journal={Tomography}, year={2021}, volume={7}, number={4}, pages={650-674}, note={Language of publication: en}, note={DOI: 10.3390/tomography7040055}, abstract={Reaction-diffusion models have been proposed for decades to capture the growth of gliomas. Nevertheless, these models require an initial condition: the tumor cell density distribution over the whole brain at diagnosis time. Several works have proposed to relate this distribution to abnormalities visible on magnetic resonance imaging (MRI). In this work, we verify these hypotheses by stereotactic histological analysis of a non-operated brain with glioblastoma using a 3D-printed slicer. Cell density maps are computed from histological slides using a deep learning approach. The density maps are then registered to a postmortem MR image and related to an MR-derived geodesic distance map to the tumor core. The relation between the edema outlines visible on T2-FLAIR MRI and the distance to the core is also investigated. Our results suggest that (i) the previously proposed exponential decrease of the tumor cell density with the distance to the core is reasonable but (ii) the edema outlines would not correspond to a cell density iso-contour and (iii) the suggested tumor cell density at these outlines is likely overestimated. These findings highlight the limitations of conventional MRI to derive glioma cell density maps and the need for other initialization methods for reaction-diffusion models to be used in clinical practice.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/334520}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/334520/1/doi\_318164.pdf}, }journal
Voxelwise principal component analysis of dynamic [s-methyl-11 c]methionine pet data in glioma patients
Martens, Corentin, Debeir, Olivier, Decaestecker, Christine, Metens, Thierry, Lebrun, Laetitia, Leurquin-Sterk, Gil, Trotta, Nicola, Goldman, Serge, and Van Simaeys, GaëtanCancers (Basel), 13, (10), 2021. DOI: 10.3390/cancers13102342.@article{info:hdl:2013/324951, author={Martens, Corentin and Debeir, Olivier and Decaestecker, Christine and Metens, Thierry and Lebrun, Laetitia and Leurquin-Sterk, Gil and Trotta, Nicola and Goldman, Serge and Van Simaeys, Ga{\"e}tan}, title={Voxelwise principal component analysis of dynamic [s-methyl-11 c]methionine pet data in glioma patients}, journal={Cancers (Basel)}, year={2021}, volume={13}, number={10}, eprint={2342}, note={SCOPUS: ar.j}, note={Language of publication: en}, note={DOI: 10.3390/cancers13102342}, abstract={Recent works have demonstrated the added value of dynamic amino acid positron emission tomography (PET) for glioma grading and genotyping, biopsy targeting, and recurrence diagnosis. However, most of these studies are based on hand-crafted qualitative or semi-quantitative features extracted from the mean time activity curve within predefined volumes. Voxelwise dynamic PET data analysis could instead provide a better insight into intra-tumor heterogeneity of gliomas. In this work, we investigate the ability of principal component analysis (PCA) to extract relevant quantitative features from a large number of motion-corrected [S-methyl-11 C]methionine ([11 C]MET) PET frames. We first demonstrate the robustness of our methodology to noise by means of numerical simulations. We then build a PCA model from dynamic [11 C]MET acquisitions of 20 glioma patients. In a distinct cohort of 13 glioma patients, we compare the parametric maps derived from our PCA model to these provided by the classical one-compartment pharmacokinetic model (1TCM). We show that our PCA model outperforms the 1TCM to distinguish characteristic dynamic uptake behaviors within the tumor while being less computationally expensive and not requiring arterial sampling. Such methodology could be valuable to assess the tumor aggressiveness locally with applications for treatment planning and response evaluation. This work further supports the added value of dynamic over static [11 C]MET PET in gliomas.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/324951}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/324951/1/doi\_308595.pdf}, }journal
2020
?A Novel Approach for Quantifying Cancer Cells Showing Hybrid Epithelial/Mesenchymal States in Large Series of Tissue Samples: Towards a New Prognostic Marker
Godin, Louis, Balsat, Cédric, Van Eycke, Yves-Remi, Allard, Justine, Royer, Claire, Remmelink, Myriam, Pastushenko, Ievgenia, D'Haene, Nicky, Blanpain, Cédric, Salmon, Isabelle, Rorive, Sandrine, and Decaestecker, ChristineCancers, 12, (4), pages 906, 2020. DOI: 10.3390/cancers12040906.@article{info:hdl:2013/304323, author={Godin, Louis and Balsat, C{\'e}dric and Van Eycke, Yves-Remi and Allard, Justine and Royer, Claire and Remmelink, Myriam and Pastushenko, Ievgenia and D'Haene, Nicky and Blanpain, C{\'e}dric and Salmon, Isabelle and Rorive, Sandrine and Decaestecker, Christine}, title={A Novel Approach for Quantifying Cancer Cells Showing Hybrid Epithelial/Mesenchymal States in Large Series of Tissue Samples: Towards a New Prognostic Marker}, journal={Cancers}, year={2020}, volume={12}, number={4}, pages={906}, note={Language of publication: en}, note={DOI: 10.3390/cancers12040906}, abstract={In cancer biology, epithelial-to-mesenchymal transition (EMT) is associated with tumorigenesis, stemness, invasion, metastasis, and resistance to therapy. Evidence of co-expression of epithelial and mesenchymal markers suggests that EMT should be a stepwise process with distinct intermediate states rather than a binary switch. In the present study, we propose a morphological approach that enables the detection and quantification of cancer cells with hybrid E/M states, i.e., which combine partially epithelial (E) and partially mesenchymal (M) states. This approach is based on a sequential immunohistochemistry technique performed on the same tissue section, the digitization of whole slides, and image processing. The aim is to extract quantitative indicators able to quantify the presence of hybrid E/M states in large series of human cancer samples and to analyze their relationship with cancer aggressiveness. As a proof of concept, we applied our methodology to a series of about a hundred urothelial carcinomas and demonstrated that the presence of cancer cells with hybrid E/M phenotypes at the time of diagnosis is strongly associated with a poor prognostic value, independently of standard clinicopathological features. Although validation on a larger case series and other cancer types is required, our data support the hybrid E/M score as a promising prognostic biomarker for carcinoma patients.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/304323}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/304323/3/GodinCancers2020.pdf}, }journal
2017
?Image processing in digital pathology: an opportunity to solve inter-batch variability of immunohistochemical staining
Van Eycke, Yves-Remi, Allard, Justine, Salmon, Isabelle, Debeir, Olivier, and Decaestecker, ChristineScientific Reports, 7, 2017. DOI: 10.1038/srep42964.@article{info:hdl:2013/246463, author={Van Eycke, Yves-Remi and Allard, Justine and Salmon, Isabelle and Debeir, Olivier and Decaestecker, Christine}, title={Image processing in digital pathology: an opportunity to solve inter-batch variability of immunohistochemical staining}, journal={Scientific Reports}, year={2017}, volume={7}, eprint={42964}, note={SCOPUS: ar.j}, note={Language of publication: en}, note={DOI: 10.1038/srep42964}, abstract={Immunohistochemistry (IHC) is a widely used technique in pathology to evidence protein expression in tissue samples. However, this staining technique is known for presenting inter-batch variations. Whole slide imaging in digital pathology offers a possibility to overcome this problem by means of image normalisation techniques. In the present paper we propose a methodology to objectively evaluate the need of image normalisation and to identify the best way to perform it. This methodology uses tissue microarray (TMA) materials and statistical analyses to evidence the possible variations occurring at colour and intensity levels as well as to evaluate the efficiency of image normalisation methods in correcting them. We applied our methodology to test different methods of image normalisation based on blind colour deconvolution that we adapted for IHC staining. These tests were carried out for different IHC experiments on different tissue types and targeting different proteins with different subcellular localisations. Our methodology enabled us to establish and to validate inter-batch normalization transforms which correct the non-relevant IHC staining variations. The normalised image series were then processed to extract coherent quantitative features characterising the IHC staining patterns.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/246463}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/246463/3/Eycke\_et\_al-2017-Scientific\_Reports.pdf}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/246463/4/supplemetary-rev-final.pdf}, }journal
2015
?An open data ecosystem for cell migration research.
Masuzzo, Paola, Martens, Lennart, 2014 Cell Migration Workshop Participants,, Ampe, Christophe, Anderson, Kurt I, Barry, Joseph, De Wever, Olivier, Debeir, Olivier, Decaestecker, Christine, Dolznig, Helmut, Friedl, Peter, Gaggioli, Cedric, Geiger, Benjamin, Goldberg, I, Horn, Ernst, Horwitz, Rick, Kam, Zvi, Le Dévédec, Sylvia SE, Vignjevic, Danijela Matic, Moore, Josh, Olivo-Marin, Jean-Christophe, Sahai, Erik, Sansone, Susanna SA, Sanz-Moreno, Victoria, Strömblad, Staffan, Swedlow, Jason, Textor, Johannes, Van Troys, Marleen, and Zantl, RomanTrends in cell biology, 25, (2), pages 55-58, 2015. DOI: 10.1016/j.tcb.2014.11.005.@article{info:hdl:2013/197899, author={Masuzzo, Paola and Martens, Lennart and 2014 Cell Migration Workshop Participants, and Ampe, Christophe and Anderson, Kurt I and Barry, Joseph and De Wever, Olivier and Debeir, Olivier and Decaestecker, Christine and Dolznig, Helmut and Friedl, Peter and Gaggioli, Cedric and Geiger, Benjamin and Goldberg, I and Horn, Ernst and Horwitz, Rick and Kam, Zvi and Le D{\'e}v{\'e}dec, Sylvia SE and Vignjevic, Danijela Matic and Moore, Josh and Olivo-Marin, Jean-Christophe and Sahai, Erik and Sansone, Susanna SA and Sanz-Moreno, Victoria and Str{\"o}mblad, Staffan and Swedlow, Jason and Textor, Johannes and Van Troys, Marleen and Zantl, Roman}, title={An open data ecosystem for cell migration research.}, journal={Trends in cell biology}, year={2015}, volume={25}, number={2}, pages={55-58}, note={SCOPUS: no.j}, note={Language of publication: en}, note={DOI: 10.1016/j.tcb.2014.11.005}, abstract={Cell migration research has recently become both a high content and a high throughput field thanks to technological, computational, and methodological advances. Simultaneously, however, urgent bioinformatics needs regarding data management, standardization, and dissemination have emerged. To address these concerns, we propose to establish an open data ecosystem for cell migration research.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/197899}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/197899/1/Elsevier\_181526.pdf}, }journal
Registration of whole immunohistochemical slide images: an efficient way to characterize biomarker colocalization.
Moles Lopez, Xavier, Barbot, Paul, Van Eycke, Yves-Remi, Verset, Laurine, Trepant, Anne-Laure, Larbanoix, Lionel, Salmon, Isabelle, and Decaestecker, ChristineJournal of the American Medical Informatics Association, 22, (1), pages 86-99, 2015. DOI: 10.1136/amiajnl-2014-002710.@article{info:hdl:2013/177042, author={Moles Lopez, Xavier and Barbot, Paul and Van Eycke, Yves-Remi and Verset, Laurine and Trepant, Anne-Laure and Larbanoix, Lionel and Salmon, Isabelle and Decaestecker, Christine}, title={Registration of whole immunohistochemical slide images: an efficient way to characterize biomarker colocalization.}, journal={Journal of the American Medical Informatics Association}, year={2015}, volume={22}, number={1}, pages={86-99}, note={JOURNAL ARTICLE}, note={SCOPUS: ar.j}, note={SCOPUS: ar.j}, note={Language of publication: en}, note={DOI: 10.1136/amiajnl-2014-002710}, abstract={Extracting accurate information from complex biological processes involved in diseases, such as cancers, requires the simultaneous targeting of multiple proteins and locating their respective expression in tissue samples. This information can be collected by imaging and registering adjacent sections from the same tissue sample and stained by immunohistochemistry (IHC). Registration accuracy should be on the scale of a few cells to enable protein colocalization to be assessed.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/177042}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/177042/7/doi\_160672.pdf}, }journal
2011
?A New Method to Address Unmet Needs for Extracting Individual Cell Migration Features from a Large Number of Cells Embedded in 3D Volumes
Adanja, Ivan, Megalizzi, Véronique, Debeir, Olivier, and Decaestecker, ChristinePloS one, 6, (7), 2011. DOI: 10.1371/journal.pone.0022263.@article{info:hdl:2013/94206, author={Adanja, Ivan and Megalizzi, V{\'e}ronique and Debeir, Olivier and Decaestecker, Christine}, title={A New Method to Address Unmet Needs for Extracting Individual Cell Migration Features from a Large Number of Cells Embedded in 3D Volumes}, journal={PloS one}, year={2011}, volume={6}, number={7}, note={SCOPUS: ar.j}, note={Language of publication: en}, note={DOI: 10.1371/journal.pone.0022263}, abstract={Background: In vitro cell observation has been widely used by biologists and pharmacologists for screening molecule-induced effects on cancer cells. Computer-assisted time-lapse microscopy enables automated live cell imaging in vitro, enabling cell behavior characterization through image analysis, in particular regarding cell migration. In this context, 3D cell assays in transparent matrix gels have been developed to provide more realistic in vitro 3D environments for monitoring cell migration (fundamentally different from cell motility behavior observed in 2D), which is related to the spread of cancer and metastases. Methodology/Principal Findings: In this paper we propose an improved automated tracking method that is designed to robustly and individually follow a large number of unlabeled cells observed under phase-contrast microscopy in 3D gels. The method automatically detects and tracks individual cells across a sequence of acquired volumes, using a template matching filtering method that in turn allows for robust detection and mean-shift tracking. The robustness of the method results from detecting and managing the cases where two cell (mean-shift) trackers converge to the same point. The resulting trajectories quantify cell migration through statistical analysis of 3D trajectory descriptors. We manually validated the method and observed efficient cell detection and a low tracking error rate (6\%). We also applied the method in a real biological experiment where the pro-migratory effects of hyaluronic acid (HA) were analyzed on brain cancer cells. Using collagen gels with increased HA proportions, we were able to evidence a dose-response effect on cell migration abilities. Conclusions/Significance: The developed method enables biomedical researchers to automatically and robustly quantify the pro- or anti-migratory effects of different experimental conditions on unlabeled cell cultures in a 3D environment. ¬© 2011 Adanja et al.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/94206}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/94206/1/journal.pone.0022263.pdf}, }journal
2005
?Tracking of migrating cells under phase-contrast video microscopy with combined mean-shift processes.
Debeir, Olivier, Van Ham, Philippe, Kiss, Robert, and Decaestecker, ChristineIEEE transactions on medical imaging, 24, (6), pages 697-711, 2005. DOI: 10.1109/TMI.2005.846851.@article{info:hdl:2013/52108, author={Debeir, Olivier and Van Ham, Philippe and Kiss, Robert and Decaestecker, Christine}, title={Tracking of migrating cells under phase-contrast video microscopy with combined mean-shift processes.}, journal={IEEE transactions on medical imaging}, year={2005}, volume={24}, number={6}, pages={697-711}, note={Evaluation Studies}, note={Journal Article}, note={Validation Studies}, note={Language of publication: en}, note={DOI: 10.1109/TMI.2005.846851}, abstract={In this paper, we propose a combination of mean-shift-based tracking processes to establish migrating cell trajectories through in vitro phase-contrast video microscopy. After a recapitulation on how the mean-shift algorithm permits efficient object tracking we describe the proposed extension and apply it to the in vitro cell tracking problem. In this application, the cells are unmarked (i.e., no fluorescent probe is used) and are observed under classical phase-contrast microscopy. By introducing an adaptive combination of several kernels, we address several problems such as variations in size and shape of the tracked objects (e.g., those occurring in the case of cell membrane extensions), the presence of incomplete (or noncontrasted) object boundaries, partially overlapping objects and object splitting (in the case of cell divisions or mitoses). Comparing the tracking results automatically obtained to those generated manually by a human expert, we tested the stability of the different algorithm parameters and their effects on the tracking results. We also show how the method is resistant to a decrease in image resolution and accidental defocusing (which may occur during long experiments, e.g., dozens of hours). Finally, we applied our methodology on cancer cell tracking and showed that cytochalasin-D significantly inhibits cell motility.}, url={http://hdl.handle.net/2013/ULB-DIPOT:oai:dipot.ulb.ac.be:2013/52108}, url={https://dipot.ulb.ac.be/dspace/bitstream/2013/52108/1/TMI2005.pdf}, }journal
Contacts
Support
- Innoviris/Région Bruxelloise
- FEDER
- FRIA/FNRS
- European Regional Development Fund
- Service Public de Wallonie Recherche
- ARC (Fédération Wallonnie - Bruxelles)
- BELSPO